Multimodal Medical AI
Contributing to GMAI-VL-R1 (RL-enhanced medical reasoning) and vision–language models for text-guided medical image generation and analysis.
Generative Medical AI · Multimodal Learning
I am a Machine Learning Researcher in the General Medical AI (GMAI) group at Shanghai AI Lab, supervised by Dr. Junjun He. My research focuses on generative AI and multimodal learning for medical applications, specializing in large-scale synthetic data generation and deep generative models. I work on developing scalable workflows to create millions of high-quality medical training samples, addressing critical challenges in data scarcity and domain adaptation for healthcare AI.
I completed my MRes with Distinction at Imperial College London (Oct 2023 - Oct 2024), supervised by Dr. Matthieu Komorowski and Dr. Guang Yang jointly. I developed deep generative models for chest X-ray image translation to improve diagnostic accuracy. During this period, I collaborated with ICU clinicians and contributed to research proposals for industrial funding.
My educational background includes a Bachelor of Science (Honours) in Data Science with University Medal from The University of Sydney, a concurrent Diploma in Computing, and a Bachelor of Science in Mathematics and Statistics from The University of Melbourne (First-Class Honours).

Portrait of a man in quest of the unknown, yet satisfied.
MRes in Machine Learning at Imperial College London. Researching generative models, synthetic data pipelines, and vision–language systems for medical imaging.
My work advances Generative AI for healthcare applications, with a focus on multimodal medical AI, large-scale synthetic data generation, and deep generative models for medical imaging.
Contributing to GMAI-VL-R1 (RL-enhanced medical reasoning) and vision–language models for text-guided medical image generation and analysis.
Building RetinaLogos-1400k (1.4M synthetic retinal images) and scalable workflows for generating millions of high-quality medical training samples.
Developing generative models for chest X-ray translation, opacity removal, and anatomical enhancement to support more accurate and robust diagnosis.
A snapshot of selected publications, academic recognition, and current goals.
4× MICCAI 2025 (1 oral, 1 spotlight) · 1× WACV 2025 · 1× IJCAI 2024 · 1× ISBI 2025 · 1× Pattern Recognition Letters · 1× NeurIPS Workshop (oral) · 3× under review (arXiv / technical reports).
I am actively seeking PhD positions for Fall 2025 and Spring 2026, as well as research internships in generative AI, multimodal learning, and medical imaging.
Feel free to reach out if you are interested in collaborations or have openings in related areas.
"Positivity is the essence of progress. In every challenge, I see an opportunity for learning and growth."
Recent work spanning generative medical imaging, multimodal reasoning, and data-centric healthcare AI.
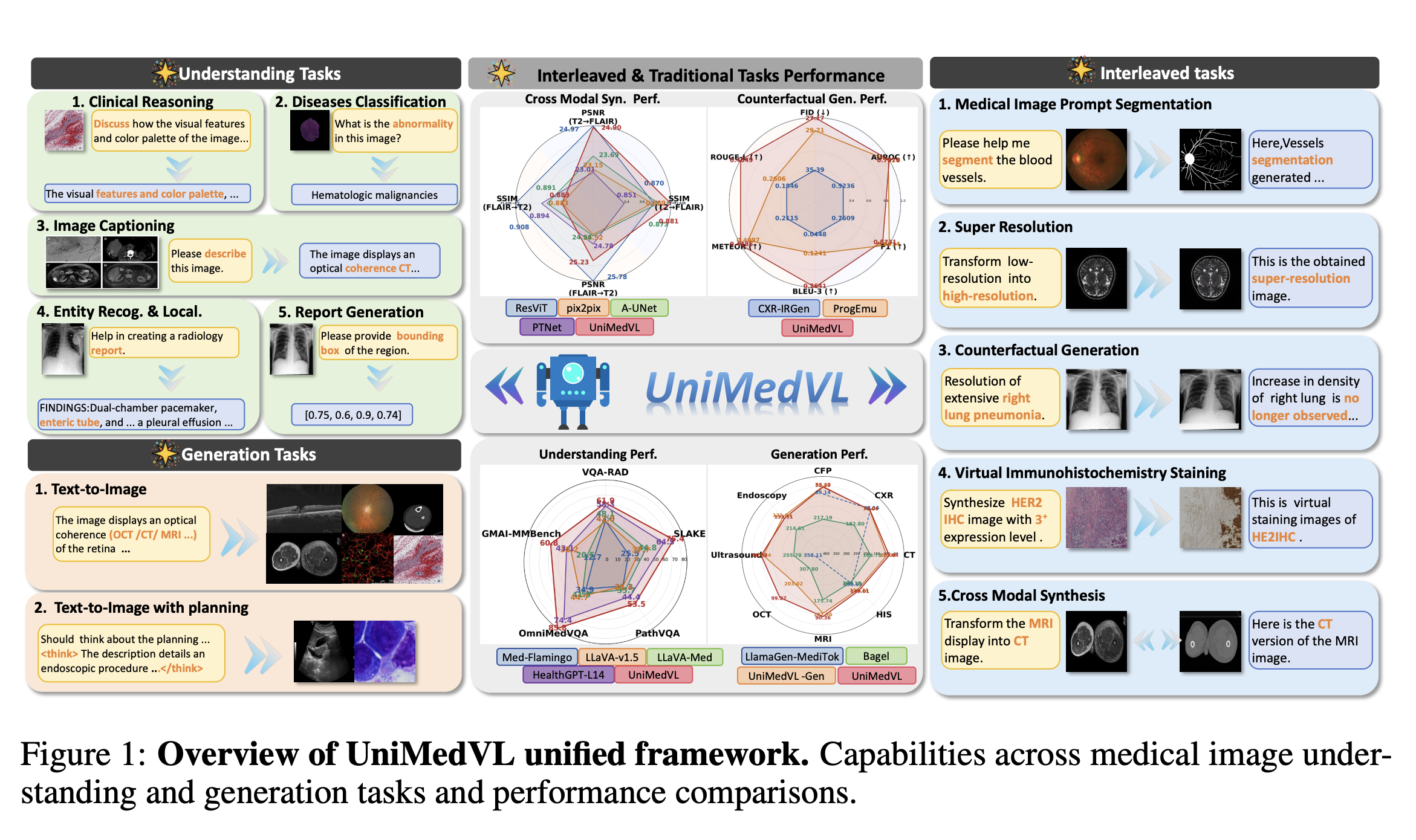
Medical diagnostic applications require models that can process multimodal inputs (images, patient histories, lab results) and generate both structured predictions and natural language explanations. We present UniMedVL, a unified medical vision-language model that bridges understanding and generation through a novel language modeling paradigm. UniMedVL achieves state-of-the-art performance on multiple medical imaging tasks including visual question answering, report generation, and diagnostic prediction while maintaining strong zero-shot generalization capabilities.
arXiv · Code · Website · Hugging Face
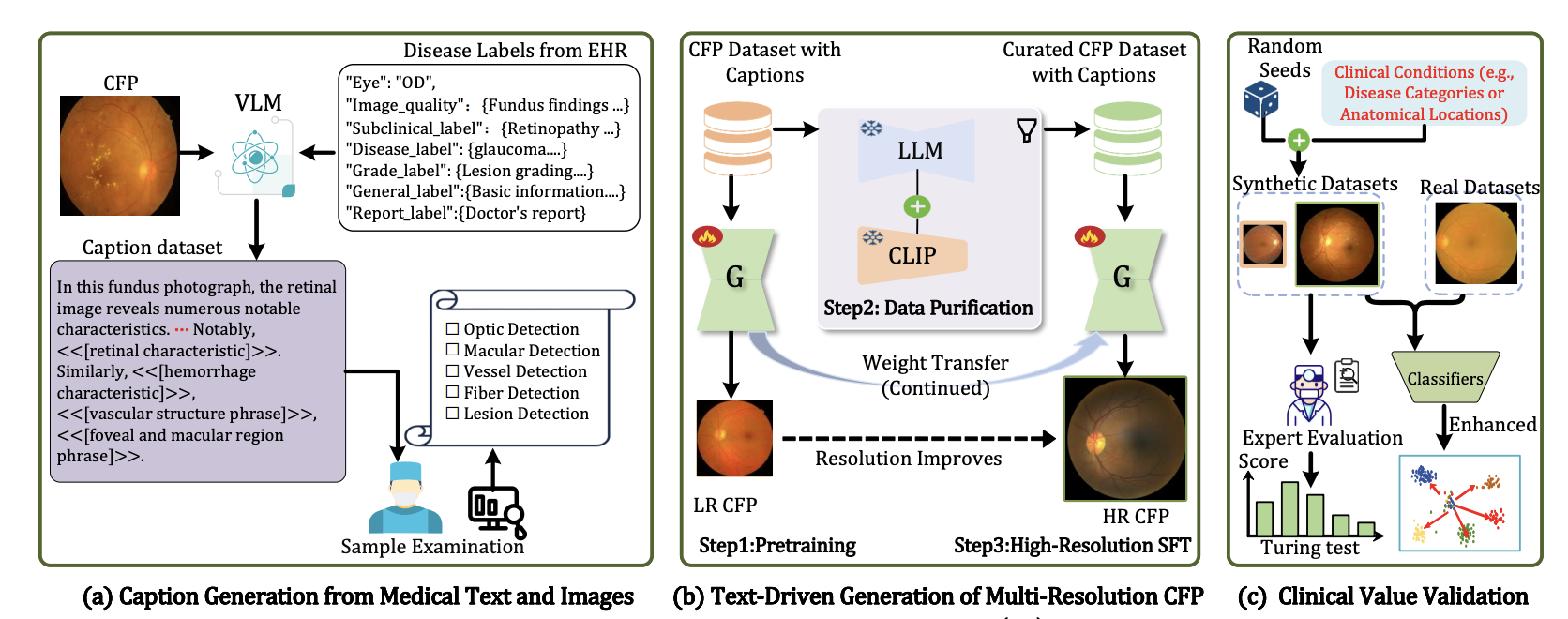
Large-scale text-to-image synthesis for retinal imaging enables unprecedented control over fine-grained anatomical features. We introduce RetinaLogos, a framework that combines latent diffusion models with the RetinaLogos-1400k dataset containing 1.4M retinal images with detailed textual descriptions. Our approach achieves superior quality in generating photorealistic retinal fundus images with precise control over pathological features, vessel structures, and optic disc characteristics, enabling robust data augmentation for downstream diagnostic tasks.
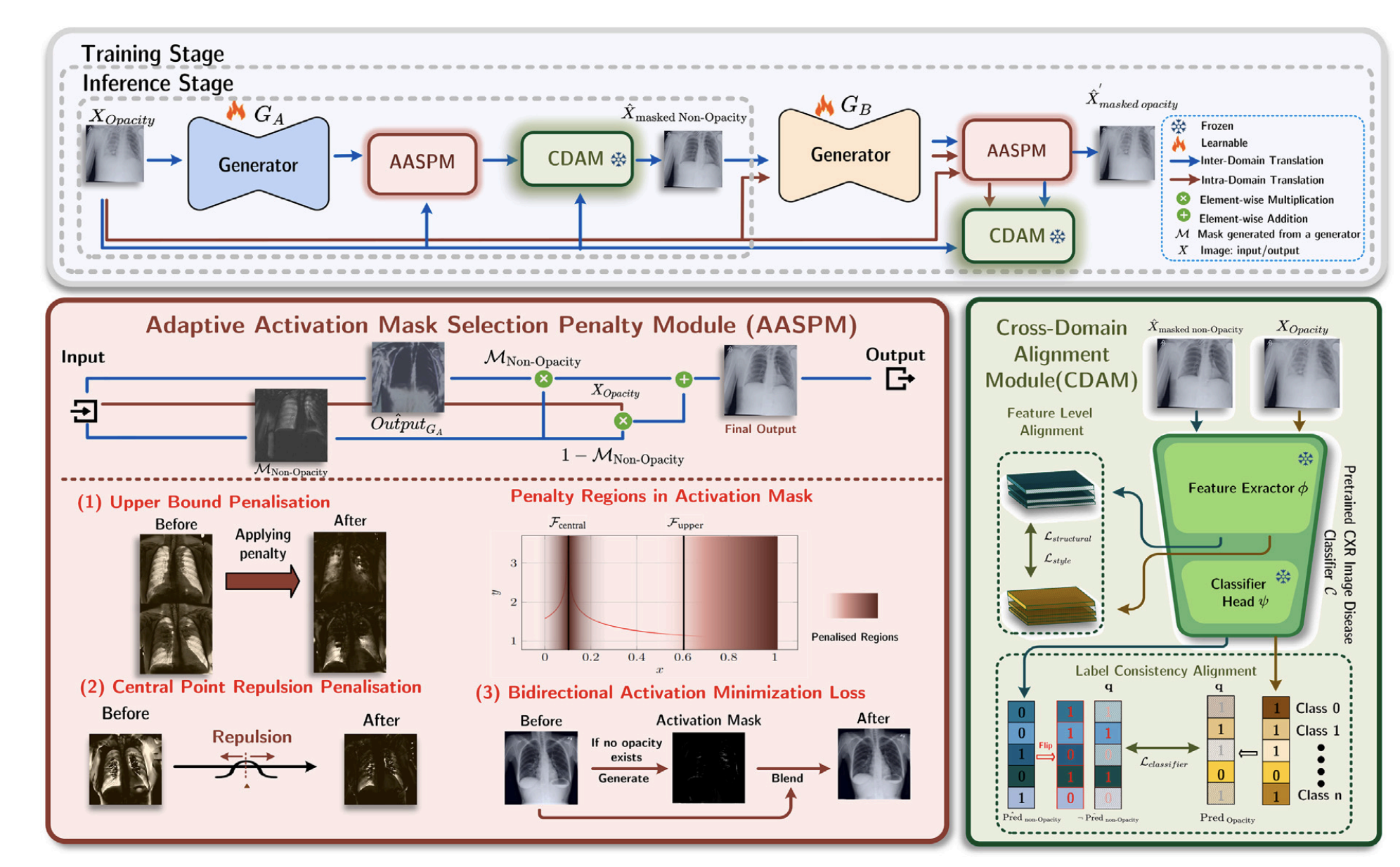
Lung opacity in chest X-rays often obscures diagnostic features, complicating disease assessment. We propose an unpaired image-to-image translation framework that removes opacity artifacts while preserving critical diagnostic content. Our method employs adaptive activation masks and cross-domain consistency constraints to learn robust mappings between normal and opacity-affected images without requiring paired training data. The approach demonstrates significant improvements in downstream segmentation accuracy and diagnostic confidence across multiple lung disease datasets.
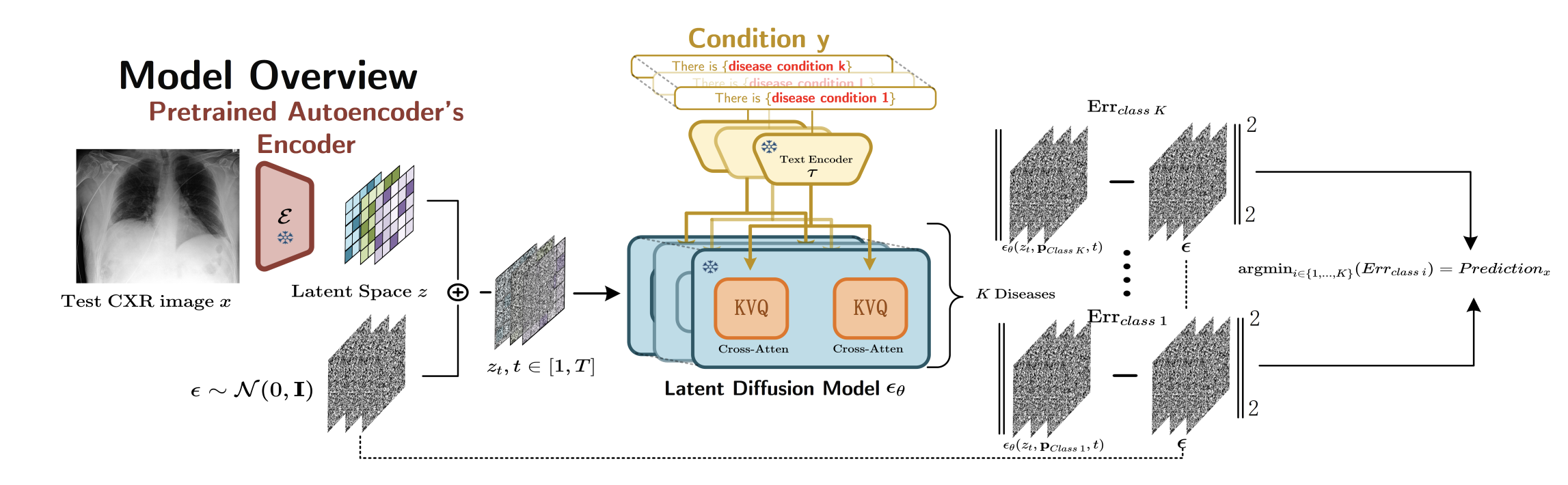
Latent diffusion models have shown remarkable performance in image generation, but their potential for medical image classification remains underexplored. We investigate how conditional latent diffusion models can be adapted for zero-shot lung disease classification in chest X-rays. Our analysis reveals that the intermediate latent representations learned during the denoising process encode rich diagnostic information, producing interpretable lesion localizations that align with radiological findings without explicit supervision for classification tasks.
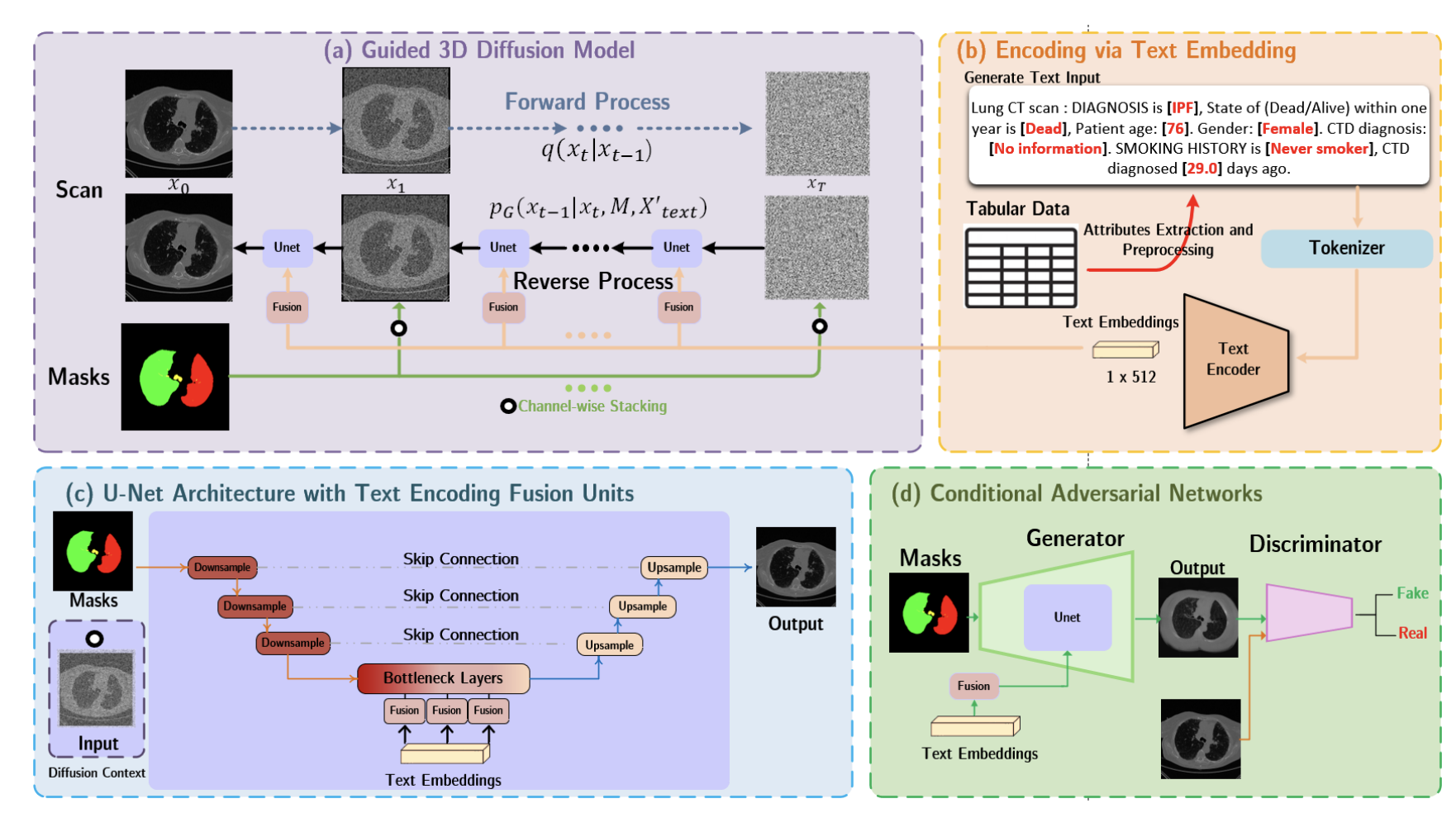
Deep generative models have revolutionized medical image analysis, but their ability to unveil underlying patterns through vision-language conditioning remains underexplored. We present a novel framework that leverages vision-language conditioning to guide deep generative models in discovering and visualizing subtle patterns in medical images. By conditioning on natural language descriptions of anatomical structures and pathological features, our approach enables interpretable pattern discovery across diverse medical imaging modalities. The framework demonstrates strong performance in revealing clinically meaningful patterns that align with radiological expertise while maintaining generative quality.
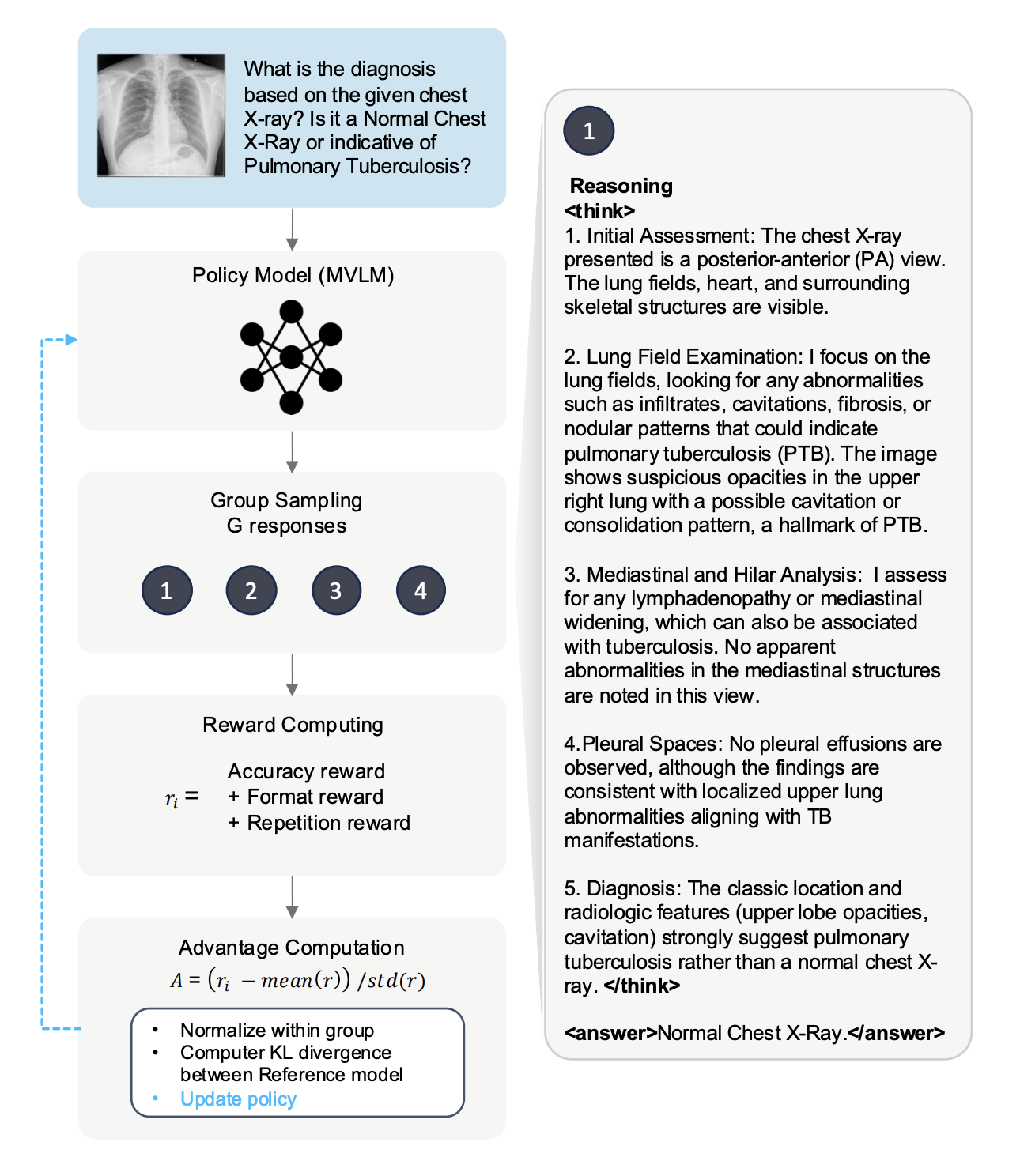
Recent advances in general medical AI have made significant strides, but existing models often lack the reasoning capabilities needed for complex medical decision-making. This paper presents GMAI-VL-R1, a multimodal medical reasoning model enhanced by reinforcement learning (RL) to improve its reasoning abilities. Through iterative training, GMAI-VL-R1 optimizes decision-making, significantly boosting diagnostic accuracy and clinical support. We also develop a reasoning data synthesis method, generating step-by-step reasoning data via rejection sampling, which further enhances the model's generalization. Experimental results show that after RL training, GMAI-VL-R1 excels in tasks such as medical image diagnosis and visual question answering.
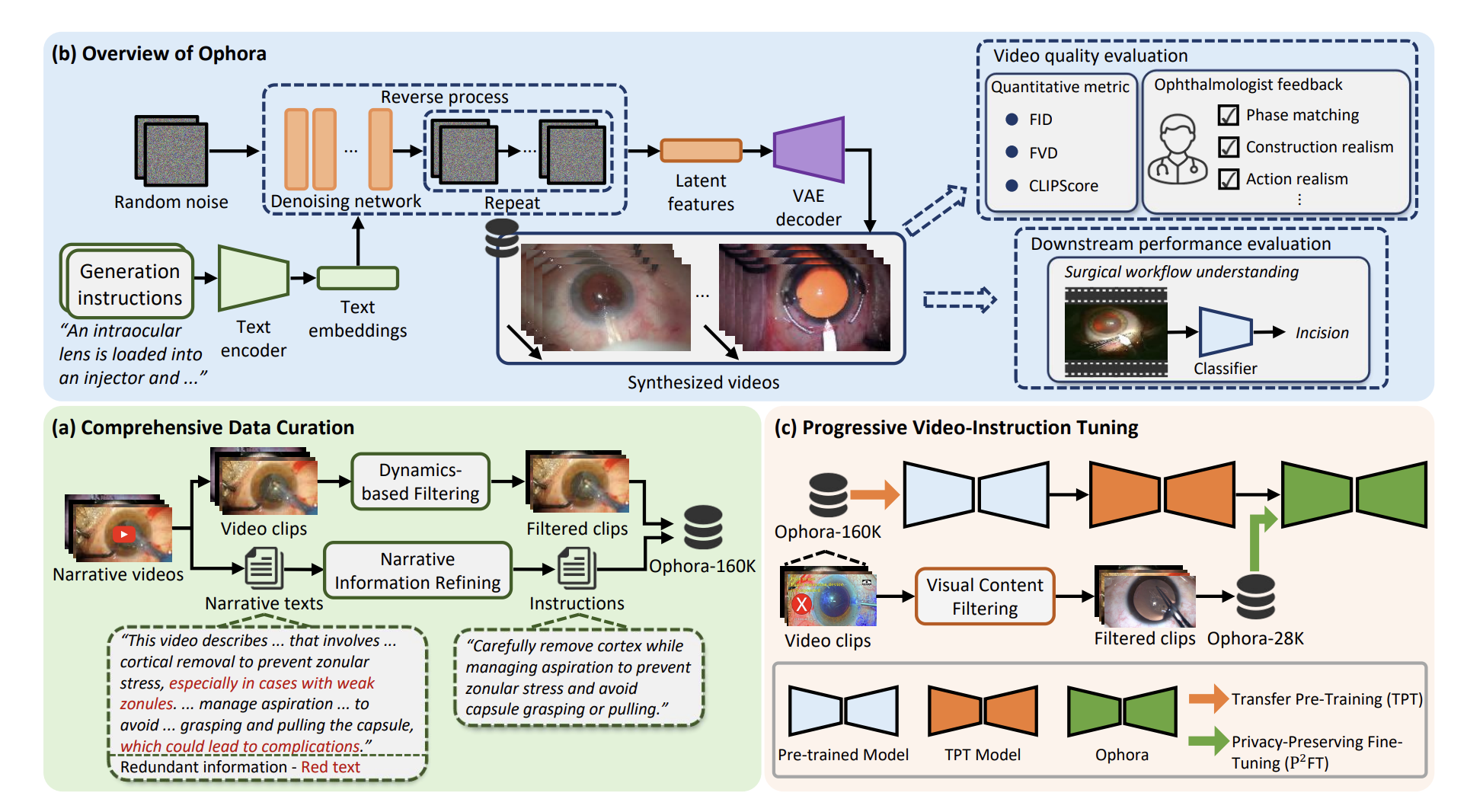
In ophthalmic surgery, developing an AI system capable of interpreting surgical videos and predicting subsequent operations requires numerous ophthalmic surgical videos with high-quality annotations, which are difficult to collect due to privacy concerns and labor consumption. Text-guided video generation (T2V) emerges as a promising solution to overcome this issue by generating ophthalmic surgical videos based on surgeon instructions. In this paper, we present Ophora, a pioneering model that can generate ophthalmic surgical videos following natural language instructions. We first propose a Comprehensive Data Curation pipeline to convert narrative ophthalmic surgical videos into a large-scale, high-quality dataset comprising over 160K video-instruction pairs, Ophora-160K.
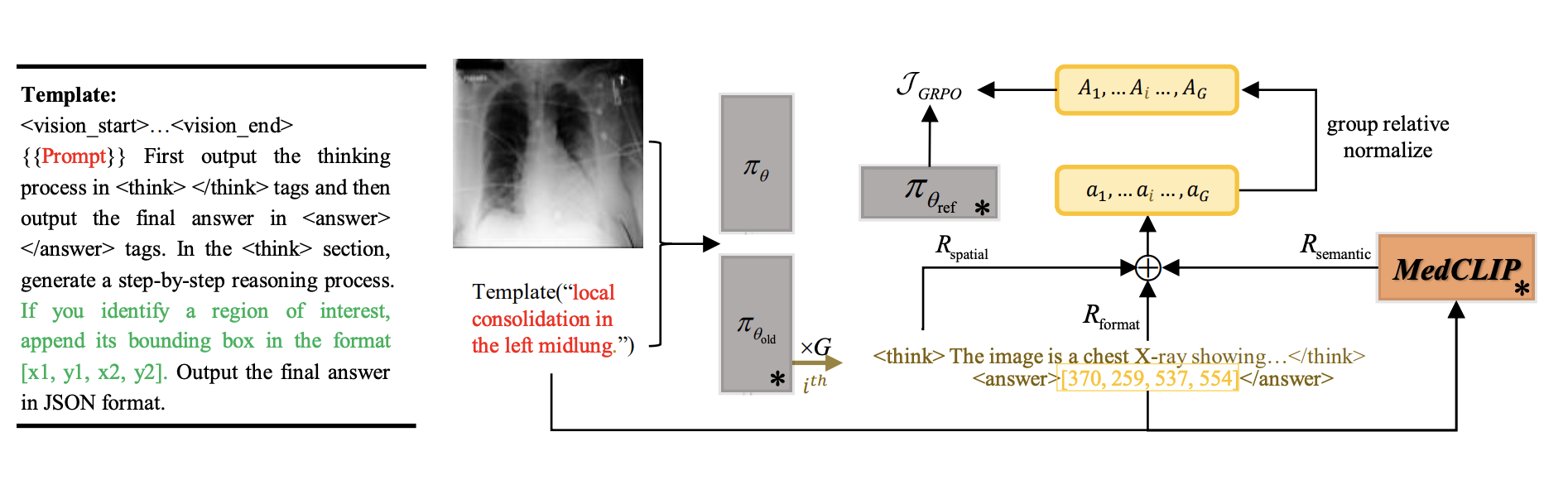
Medical Image Grounding (MIG), which involves localizing specific regions in medical images based on textual descriptions, requires models to not only perceive regions but also deduce spatial relationships of these regions. Existing Vision-Language Models (VLMs) for MIG often rely on Supervised Fine-Tuning (SFT) with large amounts of Chain-of-Thought (CoT) reasoning annotations, which are expensive and time-consuming to acquire. Recently, DeepSeek-R1 demonstrated that Large Language Models (LLMs) can acquire reasoning abilities through Group Relative Policy Optimization (GRPO) without requiring CoT annotations. In this paper, we adapt the GRPO reinforcement learning framework to VLMs for Medical Image Grounding. We propose the Spatial-Semantic Rewarded Group Relative Policy Optimization to train the model without CoT reasoning annotations.
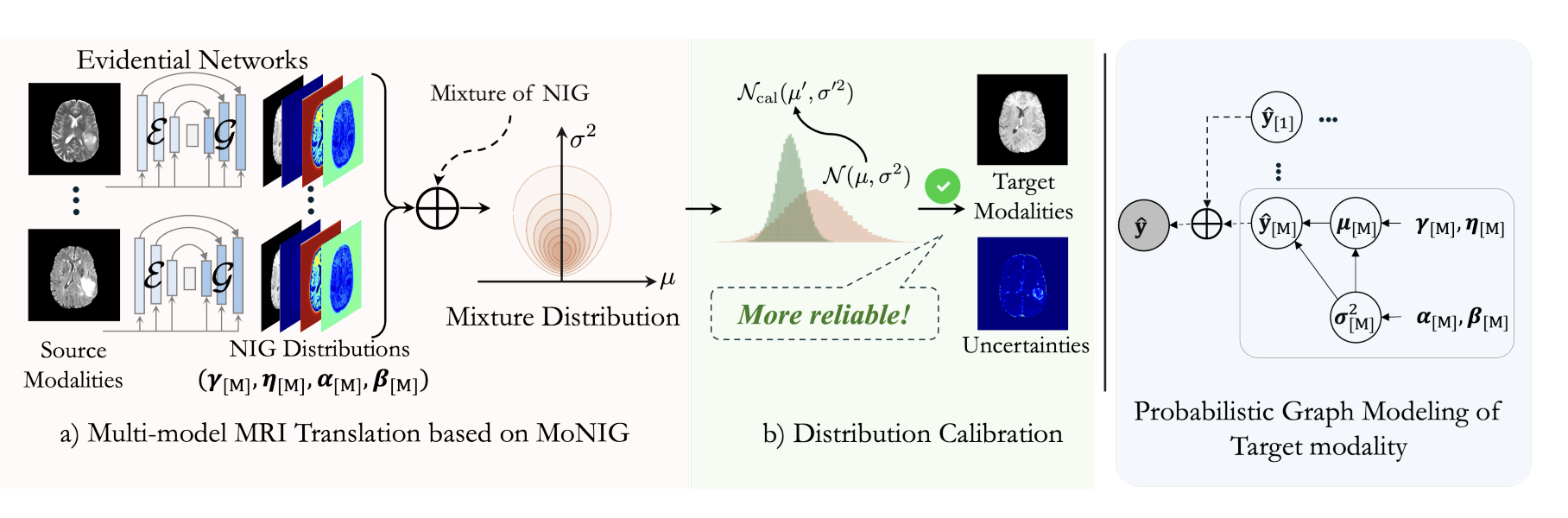
Multi-modal Magnetic Resonance Imaging (MRI) translation leverages information from source MRI sequences to generate target modalities, enabling comprehensive diagnosis while overcoming the limitations of acquiring all sequences. While existing deep-learning-based multi-modal MRI translation methods have shown promising potential, they still face two key challenges: 1) lack of reliable uncertainty quantification for synthesized images, and 2) limited robustness when deployed across different medical centers. To address these challenges, we propose a novel framework that reformulates multi-modal MRI translation as a multi-modal evidential regression problem with distribution calibration. Extensive experiments on three datasets from the BraTS2023 challenge demonstrate that our framework achieves superior performance and robustness across domains.
Paper Coming Soon
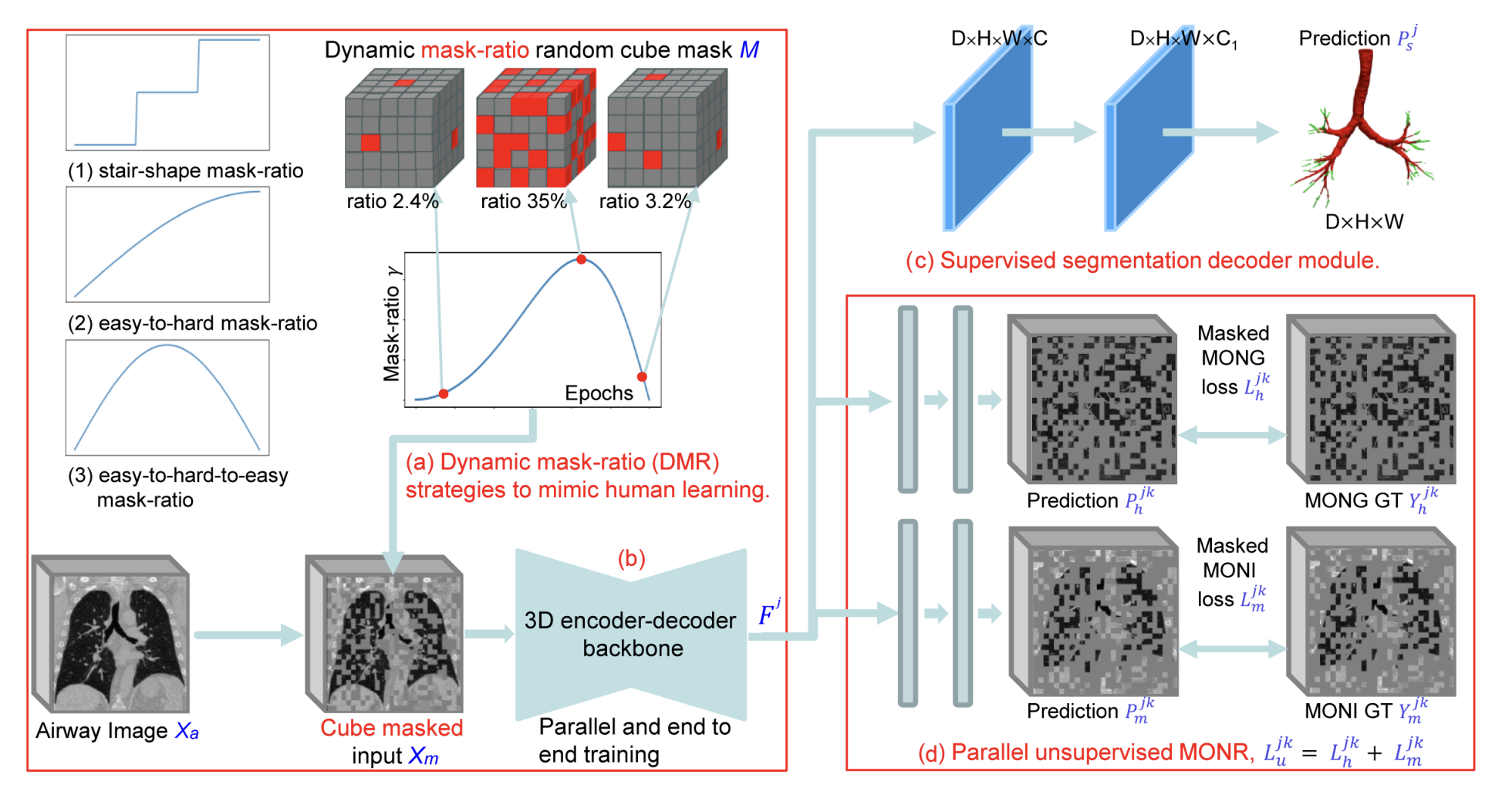
Robust airway segmentation from CT scans is challenging due to varying tree topology and imaging artifacts. DMRN introduces a dynamical multi-order response architecture that combines supervised segmentation with unsupervised structure learning. The network adaptively adjusts its receptive fields to capture both fine-grained bronchiolar details and large-scale airway topology, achieving state-of-the-art segmentation performance across diverse lung disease datasets including COPD, COVID-19, and lung cancer cohorts.
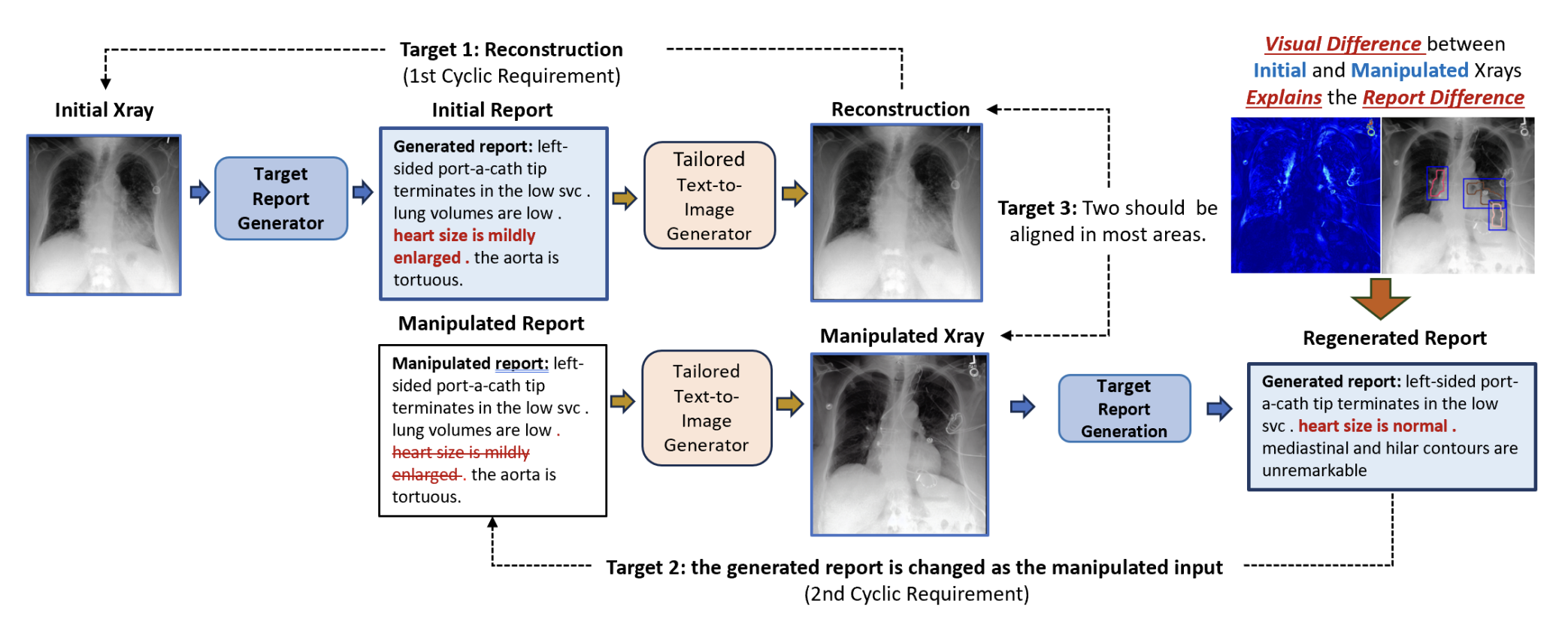
Automated medical report generation must produce reliable and interpretable outputs. We propose a cyclic manipulation framework that establishes bidirectional consistency between image features and textual reports. The model learns to manipulate visual representations in response to report modifications and vice versa, ensuring that changes in one modality produce expected changes in the other. This cyclic constraint improves both the factual accuracy and clinical reliability of generated reports while providing interpretable attention maps for key diagnostic findings.
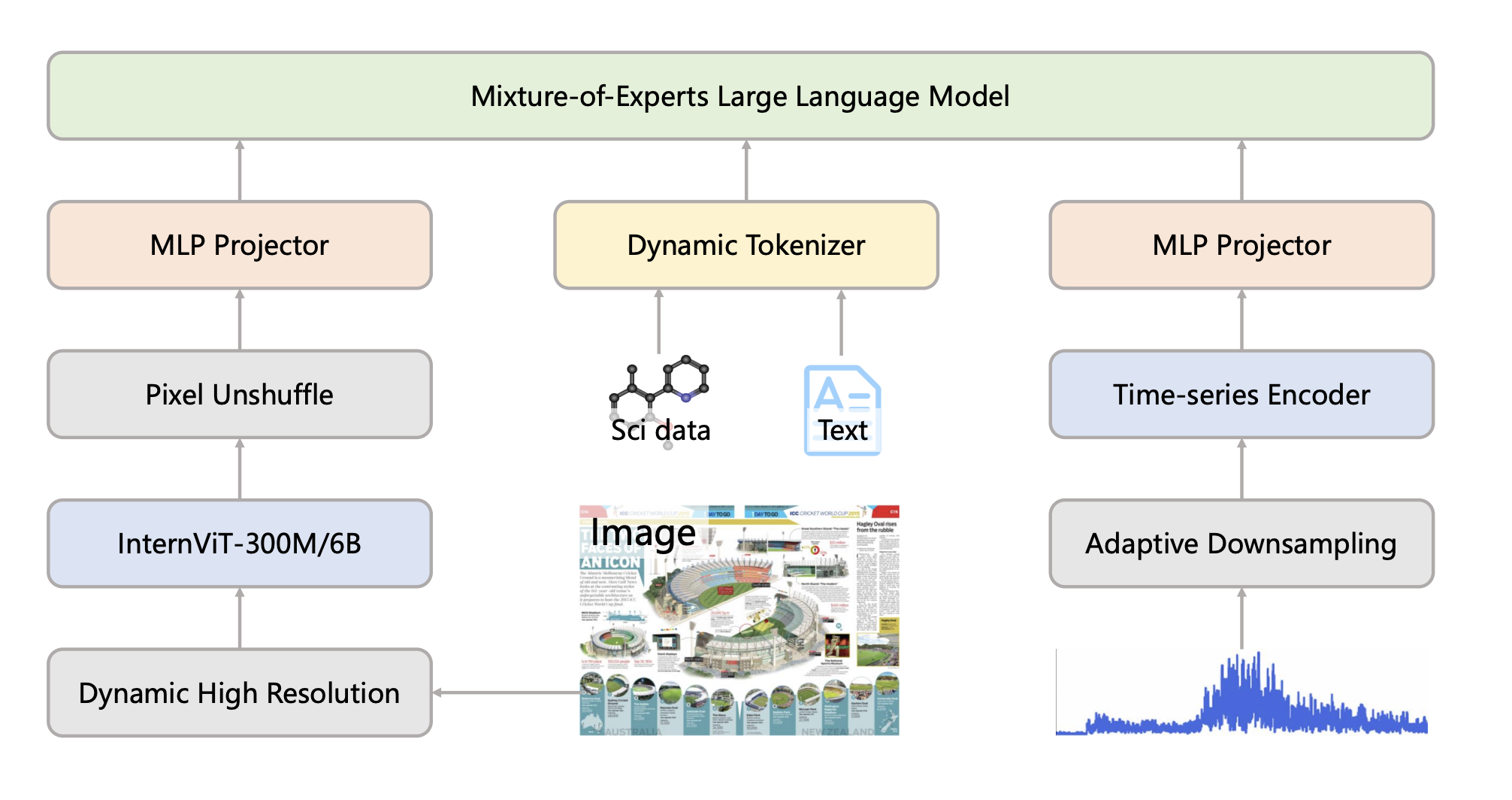
Scientific discovery requires integrating knowledge across diverse domains and modalities. Intern-S1 is a large-scale mixture-of-experts foundation model with 241B parameters, trained on scientific literature, molecular structures, experimental data, and research code. The model achieves state-of-the-art performance on molecular property prediction, crystal stability forecasting, retrosynthesis planning, and scientific question answering, demonstrating strong zero-shot transfer across chemistry, materials science, and biology.
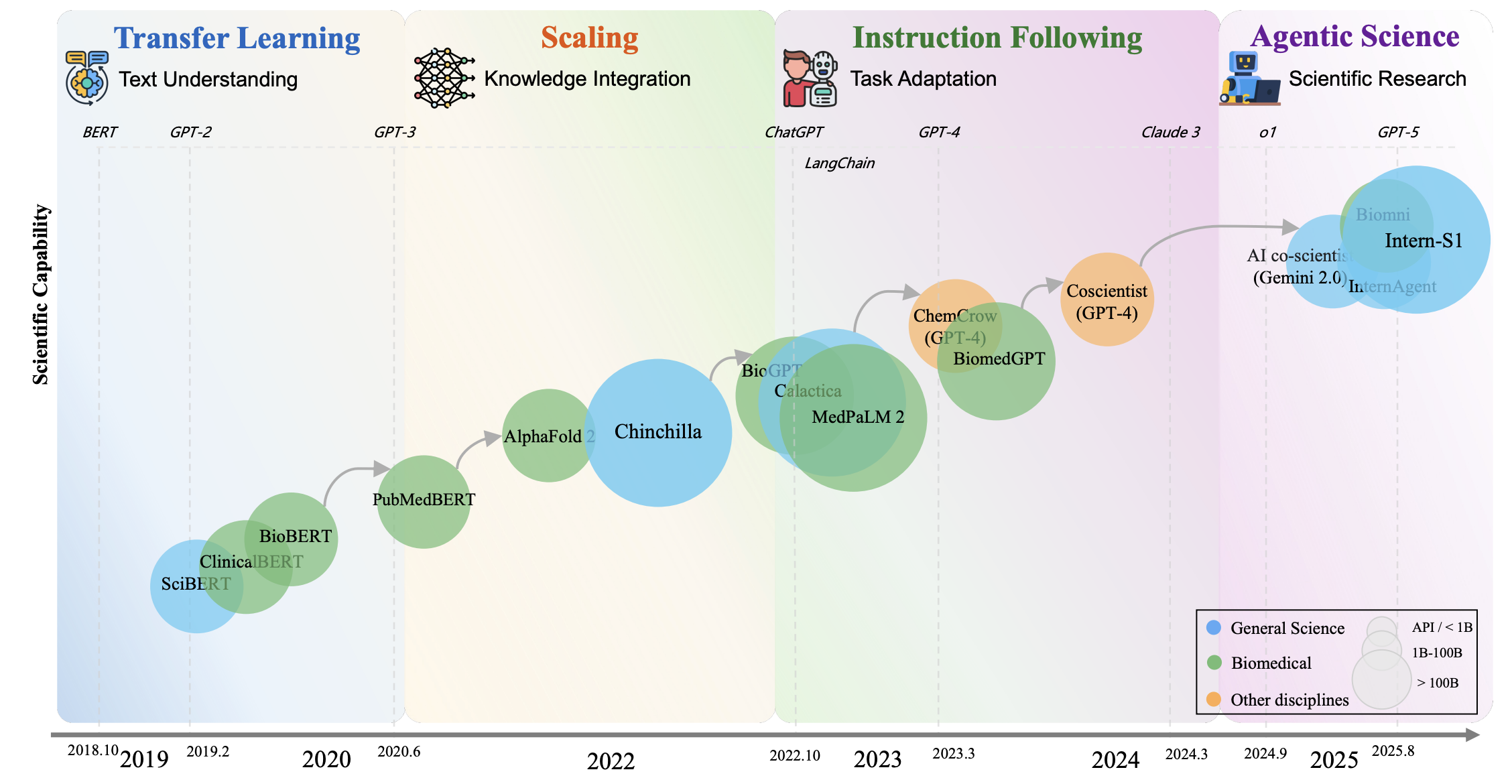
The rapid development of large language models has opened new possibilities for scientific research automation. This comprehensive survey examines scientific LLMs across the full pipeline from data curation to autonomous agents. We analyze 270+ specialized scientific datasets, 190+ domain-specific benchmarks, and emerging architectures for scientific reasoning. The survey covers applications in physics, chemistry, biology, medicine, and materials science, identifying key challenges including hallucination in scientific contexts, integration of structured scientific knowledge, and ethical considerations for AI-assisted research.
For a complete and up-to-date list of publications, please see my Semantic Scholar profile or OpenReview .
Core competencies in medical AI, generative models, and multimodal learning.
Academic achievements and recognitions throughout my research journey.
Awarded University Medal in Bachelor of Science (Honours) for achieving the highest academic distinction (WAM 89.5) in Data Science Honours program. The honour thesis on night-to-day image translation was subsequently accepted at the Australasian Database Conference.
Selected for oral presentations at MICCAI 2025 (Ophora - Ophthalmic Surgical Video Generation) and NeurIPS 2024 Workshop on Advancements in Medical Foundation Models (Deep Generative Models for Medical Imaging).
Received merit-based scholarship from University of Melbourne in recognition of academic excellence in Mathematics and Statistics (Overall WAM: 86.8/First Class Honours).
Placed on Dean's Honours List for First Year Bachelor of Science students for exceptional academic performance among the entire cohort.
A lightweight lab notebook – recent papers, positions, and project updates.
New preprint on arXiv: UniMedVL, a unified multimodal framework for medical image understanding and generation built around the Observation–Knowledge–Analysis paradigm.
4 papers accepted at MICCAI 2025, including 1 first-author paper (RetinaLogos), 1 oral presentation (Ophora), and 1 spotlight (MedGround-R1), advancing generative and multimodal medical AI.
CVLM paper on reliable and fine-grained image interpretation for automated report generation accepted at IJCAI 2025.
Unpaired chest X-ray translation method for lung opacity diagnosis accepted in Pattern Recognition Letters, improving segmentation and classification across multiple datasets.
Started as Machine Learning Researcher at Shanghai AI Lab in the GMAI group, focusing on multimodal medical AI models and large-scale synthetic dataset generation for healthcare applications.
Completed MRes (Distinction), supervised by Dr. Matthieu Komorowski and Dr. Guang Yang, working on deep generative models for chest X-ray image translation in collaboration with ICU clinicians.
From Melbourne to Sydney to London to Shanghai – a journey through data science, mathematics, and medical AI.
TMI
ICLR, CVPR, ISBI
I am deeply grateful to my supervisors Dr. Junjun He, Dr. Guang Yang, Dr. Matthieu Komorowski, and Prof. Minming Gong for their invaluable guidance, mentorship, and support throughout my research journey. Their insights and encouragement have been instrumental in shaping my academic growth.
I also extend my sincere thanks to my collaborators Lihao Liu, Sheng Zhang, Xiaodan Xing, Yingying Fang, Cheng Tang, Wei Li, Jiyao Liu, Huihui Xu, and many others. Their expertise, dedication, and teamwork have been essential to our research achievements.
I am always happy to chat about generative models, multimodal systems, and new collaborations in medical AI.
If you are working on generative medical AI, multimodal learning, or data-centric healthcare, I would love to connect.